YesWorkflow is a set of software tools that aims to provide users of scripting languages (such as Perl, Python, R, and MATLAB) with many of the benefits of scientific workflow systems such as Kepler. YesWorkflow requires neither the use of a workflow engine nor the overhead of adapting code to run e lectively in such a system. Instead, YesWorkflow enables scientists to annotate existing scripts with special comments that reveal the computational modules and dataflows otherwise implicit in these scripts. YesWorkflow tools extract and analyze these comments, represent the scripts in terms of entities based on the typical scientific workflow model, and provide graphical renderings of this workflow-like view of the scripts. Future versions of YesWorkflow will also allow the prospective provenance of the data products of these scripts to be queried in ways similar to those available to users of scientific workflow systems.
The current revision and technical details on YesWorkflow technical details see our Github YesWorkflow Site.
McPhillips, Timothy, Tianhong Song, Tyler Kolisnik, Steve Aulenbach, Khalid Belhajjame, Kyle Bocinsky, Yang Cao, James Cheney, Fernando Chirigati, Saumen Dey, Juliana Freire, Christopher Jones, James Hanken, Keith W. Kintigh, Timothy A. Kohler, David Koop, James A. Macklin, Paolo Missier, Mark Schildhauer, Christopher Schwalm, Yaxing Wei, Mark Bieda, Bertram Ludäscher. 2015 YesWorkflow: A User-Oriented, Language-Independent Tool for Recovering Workflow Information from Scripts. International Journal of Digital Curation 10(1): 298–313. DOI: 10.2218/ijdc.v10i1.370
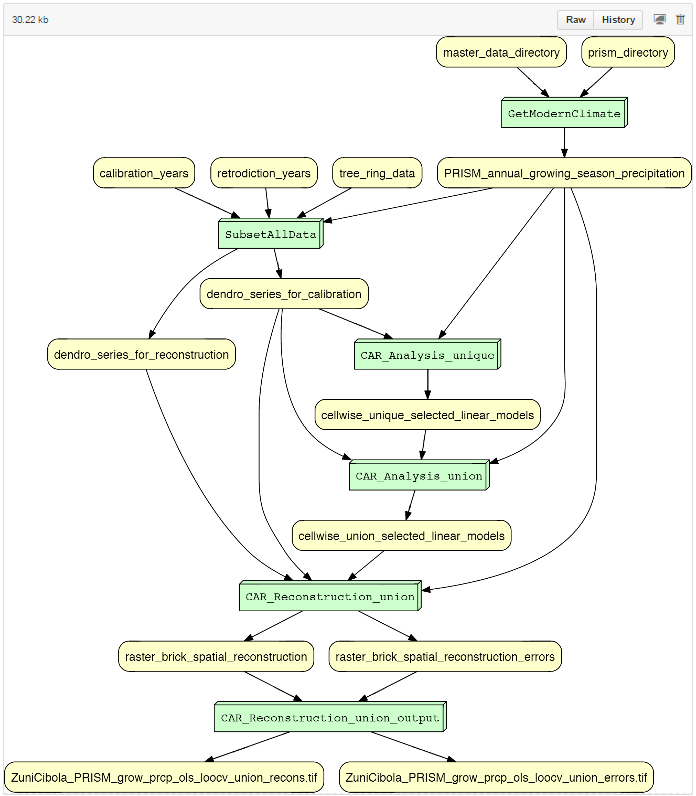
Below is an example of YesWorkflow output for the PaleoCAR environmental reconstruction process implemented in R: